Google DeepMind has identified over two million crystal structures with its GNoME artificial intelligence (AI) tool as it follows up its protein synthesis efforts in recent years. If proven, these crystal substances could be tested in energy applications, including being the raw materials for solar cells and special electrical devices with zero resistance at certain temperatures.
Of the two million-odd substances DeepMind has discovered, they plan to make 381,000 available for further testing. The AI findings are equivalent to the knowledge previously gained through 800 years of testing and experiments.
DeepMind AI Crystal Research Lowers Entry Barrier
However, the team needed human insight to gauge the stability of the AI chemical structures. In a paper published in Nature, the scientists said AI helped bypass “expensive trial-and-error” methods.
Read more: Will AI Replace Humans?
Their findings are already being used by researchers at the University of California, Berkeley, and the Lawrence Berkeley National Laboratory. In addition to solar and electrical applications, the crystal structures can be used to create computer chips for big companies like Nvidia and IBM. American electrical engineer Jack Kilby built the first computer chip on a silicon crystal in 1958.
DeepMind previously used AI to create protein structures with atom-level accuracy. Biotechnology companies are using the technology for vaccines and other drug discovery experiments.
Read more: The 6 Hottest Artificial Intelligence (AI) Jobs in 2023
The technology may help bypass the expensive process of laboratory synthesis, although the drugs still need to undergo clinical trials. Three-dimensional protein prediction was first proposed by C. B. Anfinsen, E. Haber, M. Sela, and F. H. White, Jr. in a 1961 National Academy of Sciences paper.
But Can DeepMind Solve Hard Problems?
The discovery of new crystal structures and proteins are important scientific breakthroughs. But as one chemistry expert, Derek Lowe, points out, DeepMind’s protein tool cannot make sense of less-known protein regions that do not have an ordered structure. He contends that knowing a protein structure is not the main challenge when making drugs.
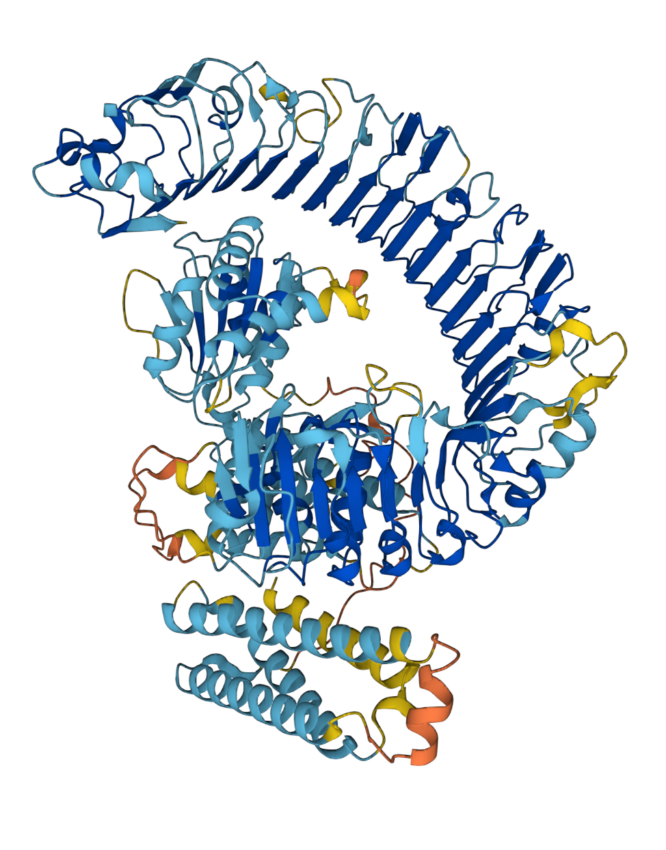
“The protein’s structure might help generate ideas about what compounds to make next, but then again, it might not. In the end, the real numbers from the real biological system are what matter.”
He adds that drugs are more likely to fail because researchers don’t use them used correctly or if they behave in odd ways. An understanding of the protein structure does little to mitigate these risks.
Do you have something to say about crystal or protein structures developed by AI company DeepMind, or anything else? Please write to us or join the discussion on our Telegram channel. You can also catch us on TikTok, Facebook, or X (Twitter).
Disclaimer
In adherence to the Trust Project guidelines, BeInCrypto is committed to unbiased, transparent reporting. This news article aims to provide accurate, timely information. However, readers are advised to verify facts independently and consult with a professional before making any decisions based on this content. Please note that our Terms and Conditions, Privacy Policy, and Disclaimers have been updated.


